FIG

.
Technology
SEED
The SEED Project was started in 2003 by the
Fellowship for Interpretation of Genomes
(FIG) as a largely unfunded open source
effort. Argonne National Laboratory,and the
University of Chicago joined the project in
2004, and since then much of the activity
occurred at those two institutions. Other
active participants came form the University
of Illinois at Urbana-Champaign, Hope
college, San Diego State University, the
Burnham Institute and a number of other
institutions. The cooperative effort focused
on the development of the comparative
genomics and annotation environment called
the SEED and, more importantly, on the
development of curated genomic data.
The Pathosystems Resource Integration Center is one of four bioinformatics resource centers (BRCs) funded by the National Institute of Allergy and Infectious Diseases (NIAID). The BRC program supports research by providing access to data associated with the NIAID Category A-C pathogenic genera, with PATRIC serving as the bacterial database. In order to provide a rich comparative analysis environment, PATRIC also provides access to all publicly available genomes and associated metadata for bacterial and archaeal isolates, which includes more than 100,000 genomes.
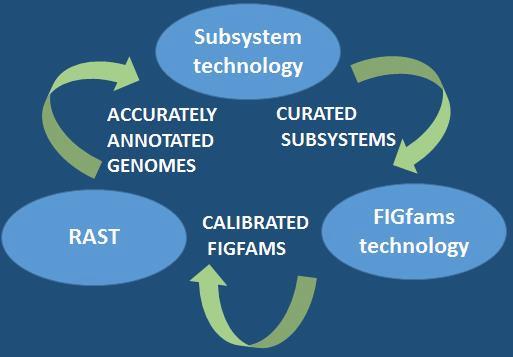
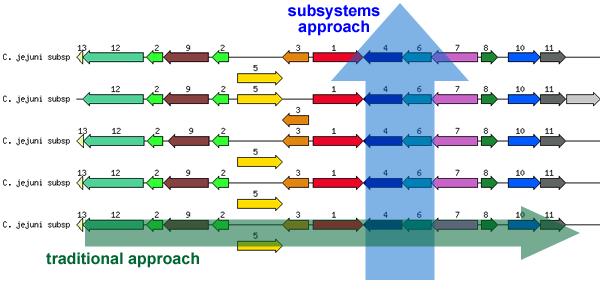
Subsystems are co-curated sets of biologically related functional roles across all genomes. They are units of expert knowledge about a single biological process like a metabolic pathway, (or pieces of a pathway), subunits of multi-unit enzymes, or a cellular structure such as the Ribosome and more. Subsystems allow the annotator to curate homologous functional roles across all genomes simultaneously (blue vertical arrow in the diagram below), rather than annotating one genome at a time jumping from gene to gene, without having any specific knowledge about most of the genes (green horizontal arrow in the diagram). By studying the literature about a given subsystem and curating the respective functional roles, the annotator rapidly develops expertise in the variations, which are present in different organisms. This enables faster, more accurate annotations and allows to recognize missing pieces of biochemistry or new variations of pathways in new genomestions, which are present in different organisms. This enables faster, more accurate annotations and allows to recognize missing pieces of biochemistry or new variations.
Subsystems
RAST is an automatic annotation server for microbial genomes, built upon the framework provided by the SEED system. More then 100,000 genomes have been processed by RAST since its deployment in 2007. RAST is designed to consistently produce annotations comparable in quality to those produced by human annotators and to extend those annotations to as many protein-encoding genes in as many genomes as possible. Continuous addition of new subsystems that cover previously un-annotated regions of the genomes, and continuous quality control of existing subsystems are central to improved annotations in the SEED and their propagation via FIGfams (and now PATtyFams) into RAST. RAST is the annotation engine of PATRIC.
FIGfams are subsystem-derived protein families of isofunctional homologs. They are thought of as implementing one or more abstract functional roles. All members of a single FIGFam are believed to implement precisely this same set of functional roles. The original set of FIGfams has been supplanted by the PATtyFams .
FIGFams